
SARS-CoV-2 Multiplex
Qualitative RT-PCR-Based Detection of SARS-CoV-2 For in vitro diagnostic use. For professional use only.
![]()

Intended Use and Test Principle
This document describes the use of real-time RT-PCR assays for the in vitro qualitative detection of 2019-Novel Coronavirus (SARS-CoV-2), which causes COVID-19, in respiratory specimens. The SARS-CoV-2 primer and probe sets are designed for the specific detection of SARS-CoV-2.
Diagnovital SARS-CoV-2 Multiplex Real-Time PCR Kit is an in vitro nucleic acid amplification assay for qualitative detection of 2019-Novel Coronavirus (SARS-CoV-2) in respiratory specimens using Viral Nucleic Acid Isolation Kit and BIO-RAD CFX96-IVD or Rotor-Gene 3000/6000 or Himedia Insta Q96 or Applied Biosystems 7500 or QuantStudio 5 Real-Time PCR Detection Systems for amplification, detection and analysis.
The kits follow CDC’s and WHO’s latest detection guidelines.
Product Description
Diagnovital SARS-CoV-2 Multiplex is a real-time RT-PCR-based detection system for the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is considered a novel human coronavirus that is genetically distinct from the common human coronaviruses (229E, NL63, OC43, HKU1), which cause seasonal acute respiratory illness. It is also genetically distinct from the two newer human coronaviruses, MERS-CoV and SARS-CoV.
Diagnovital SARS-CoV-2 Multiplex detects the presence of 2 different and highly specific gene sequences of corona viruses (N1 and ORF1ab) at the FAM channel and one sequence specific for human RNA serving as a human extraction control (HEC, RNaseP) at the HEX/VIC channel. All 2 assays must be tested positive to confirm the sample as SARS-CoV-2-positive.
Additionally, a non-infectious positive control and a negative human extraction control are included. Human Extraction Control (HEC) is needed to ensure appropriate RNA extraction, purification and reverse transcription and all reagents involved in reaction. The Human Extraction Control (HEC) master mix contains primers and probe for an endogenous human target, which is extracted from the swab during the extraction step. We do not put an external DNA or RNA template as extraction control, since we already get human target during extraction. The positive control is used to confirm functionality of the assays and overall PCR performance, the negative human extraction control is included to evaluate the quality of the RNA isolation independently from the SARS-CoV-2 assays.
Real Time PCR-Based Detection of SARS-CoV-2
The first step in the detection of SARS-CoV-2 is the conversion of viral RNA into cDNA. Afterwards, the target sequences unique for SARS-CoV-2 and HEC are specifically amplified with amplification monitored in real time through the use of fluorescently labelled probes: upon incorporation into the newly amplified DNA strands, the fluorophore is released and an increase in fluorescence signal can be observed.
Due to the intrinsic mutation rate of coronaviruses, it is possible that mutations in the target sequence occur and accumulate over time. This can lead to false-negative results with a PCR-based detection approach. DIAGNOVITAL® SARS-CoV-2 Multiplex addresses this issue by using 2 detection assays on 2 different target sequences (N1 and ORF1ab) to minimize the chance of false-negative results caused by an altered target sequence.
If samples are tested negative in one or more assays, additional complementary testing may be required. The original target sequences for SARS-CoV-2 are included as a non-infectious target positive control (TPC) to check the integrity of the detection assays.
Samples tested positive should always be confirmed through complementary methods and additional analysis in an independent laboratory.
DIAGNOVITAL® SARS-CoV-2 Multiplex is compatible with every qPCR with calibrated FAM™ and HEX/VIC channel, whereas normalization with the ROX channel is optional.
Materials Provided
| Reagents | Quantity and Volume (25 tests) | Quantity and Volume (50 tests) | Quantity and Volume (100 tests) | |
| 1 | 50X VitaScriptTM Reverse Transcriptase | 1 × 25 µl | 1 × 50 μl | 1 × 100 μl |
| 2 | Diagnovital® 2X SARS-CoV-2 Multiplex Mix | 1 × 375 µl | 1 × 750 μl | 1 × 1500 μl |
| 3 | SARS-CoV-2 Target Positive Control (TPC) | 1 × 45 µl | 1 × 75 μl | 1 × 150 μl |
| 4 | Nuclease-free dH2O | 1 × 1000 μl | 1 × 1000 μl | 1 × 1000 μl |
Additional Materials Required
- Suitable means & equipment for nucleic acid extraction
- Real-time PCR detection system equipped for FAMTM and HEX/VIC detection
- Adjustable pipettes & fitting filtered pipette tips
- Appropriate personal protective equipment & workspaces for working with potentially infectious samples
- Surface decontaminants such as DNAZapTM (Life Technologies), DNA AwayTM (Fisher Scientific), RNAse AwayTM (Fisher Scientific), 10% bleach (1:10 dilution of commercial 5.25-6.0% sodium hypochlorite)
- Nuclease-free tubes / strips / plates to prepare dilutions, master mixes etc.
- Nuclease-free tubes / strips / plates corresponding to the PCR device
- Suitable storage options for reagents and specimen (4°C, -20°C, -70°C)
Storage
- Store all components at -20°C and avoid repeated freeze and thaw cycles.
- Protect the 2X qPCR master mixes from light as prolonged exposure can diminish the performance of the fluorophores.
- If the kit components have been damaged during transport, contact supplier immediately. Do not use as performance may be compromised.
- Keep reagents separate from sample material to avoid contamination.
- Do not use after the designated expiry date.
Performance Characteristics
Analytical sensitivity
Analytical sensitivity was analyzed by use of a dilution series of DIAGNOVITAL® SARS-CoV-2 Multiplex Reference samples. A dilution series of a DIAGNOVITAL® SARS-CoV-2 Multiplex Reference samples was prepared to give the final concentrations of 300, 100, 30 and 10 copies/ml. Each dilution was tested in 24 replicates. Lower limit was calculated by probit analysis done by PASW Statistics 18 program. For each genotype/subtype, Limit of Detection (LoD) values and 95% confidence ranges are summarized in Table 1.
| Target Gene | Limit of Detection (copies/mL) | 95% Confidence Lower Limit | 95% Confidence Upper Limit |
| N1+ORF1ab | 38 | 33 | 50 |
Table 1: DIAGNOVITAL® SARS-CoV-2 Multiplex PCR Kit - Limit of Detection (LoD) values and 95% confidence ranges
Precision
In this study, precision of the kit was evaluated for intra-assay, inter-assay, inter-batch, by using Viral NA Isolation Kit (Cat No: 09029100) and different specimen types (oropharyngeal vs. nasopharyngeal swabs). For each target gene and different assay, 24 replicates of 103 copies/ml DIAGNOVITAL® SARS-CoV-2 Multiplex Reference samples were used. Descriptive statistics were analyzed by IBM SPSS Statistics program. Overall precision assays associated with Ct values were summarized in Table 2.
| Descriptive Statistics | |||||
| Target Gene | N | Mean | Std. Deviation | Variance | Coefficient of Variation (%) |
| N1+ORF1ab | 96 | 23.6175 | 0.139198 | 0.019269875 | 0.595032 |
Table 2: Overall descriptive statistics of DIAGNOVITAL® SARS-CoV-2 Multiplex precision data
Diagnostic specificity
SARS-CoV-2 RNA negative clinical specimens were analyzed to determine the diagnostic specificity of DIAGNOVITAL®.
DIAGNOVITAL® SARS-CoV-2 Multiplex Real Time PCR Kit. 30 SARS-CoV-2 RNA negative clinical oropharyngeal swab specimens and 30 SARS-CoV-2 RNA negative clinical oropharyngeal swab specimens and 30 bronchoalveolar lavage specimens were used. None of the 100 SARS-CoV-2 negative clinical specimens gave positive test result for SARS-CoV-2. Diagnostic specificity of DIAGNOVITAL® SARS-CoV-2 Multiplex Real Time PCR Kit is 100%. All of the Human Extraction Controls (HEC) of tests gave positive result.
Cross-Reactivity
To examine the specificity of an assay, cross-reactivity studies should be performed for potential cross-reactive markers. In this study, the specificity of the assay was evaluated by testing. 20 reference organisms.
DIAGNOVITAL® SARS-CoV-2 Multiplex Real Time PCR Kit do not show any cross-reactivity with other potential cross-reactive markers given in the table 3 below:
| Sample | Source |
| Human Adenovirus | NIBSC (Cat. No: 16/324) |
| Parainfluenza virus | ATCC VR-93 |
| Influenza A | ATCC VR-95 |
| Influenza A H5N1 | ATCC VR-1609 |
| Influenza A H1N1 | ATCC VR-1672 |
| Influenza A H3N2 | ATCC VR-822 |
| Influenza A H7N7 | ATCC VR-1641 |
| Influenza B | ATCC VR-101 |
| Parainfluenza 1 | ATCC VR-94 |
| Parainfluenza 2 | ATCC VR-92 |
| Parainfluenza 3 | ATCC VR-93 |
| Parainfluenza 4 | ATCC VR-579 |
| Human Metapneumovirus (hMPV) | ATCC VR-3250SD |
| Human Enterovirus V71 | ATCC VR-1432 |
| Human respiratory syncytial virus | ATCC VR-154 |
| Human Coronavirus NL63 | ATCC VR-3263SD |
| Human Coronavirus HKU1 | ATCC VR-3262SD |
| Human Coronavirus 229E | ATCC VR-740 |
| Betacoronavirus 1 OC43 | ATCC VR-1558D |
| MERS Coronavirus | ATCC VR-3248SD |
Table 3: Potential cross-reactive markers tested in the study
Considerations Before Starting
Bıosafety
- Wear appropriate personal protective equipment (e.g. gowns, powder-free gloves, eye protection) when working with clinical specimen.
- Specimen processing should be performed in a certified class II biological safety cabinet following biosafety level 2 or higher guidelines.
- For more information, refer to: Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Patients Under Investigation (PUIs) for 2019 Novel Coronavirus (SARS-CoV-2).
https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html
- Biosafety in Microbiological and Biomedical Laboratories 5th Edition available at
http://www.cdc.gov/biosafety/publications
- The use of DIAGNOVITAL® SARS-CoV-2 Multiplex is restricted to trained laboratory personnel only.
Specimens
Only use appropriate specimens for testing, such as:
- Respiratory specimens including nasopharyngeal / oropharyngeal swabs and bronchoalveolar lavage.
- Swab specimens should be collected only on swabs with a synthetic tip (such as polyester or Dacron®) with plastic shafts. Swabs with calcium alginate or cotton tips with wooden shafts are not acceptable.
Specimens – Handling and Storage
- Specimens can be stored at 4°C for up to 72 hours after collection.
- If a delay in extraction is expected, store specimens at -70°C or lower.
- Extracted nucleic acids should be stored at -70°C or lower.
Do not use specimens if:
- They were not kept at 2-4°C (≤ 4 days) or frozen at -70°C or below.
- They are insufficiently labelled or lack documentation.
- They are not suitable for this purpose (see above for suitable sample material).
- The specimen volume is insufficient.
Sample Preparation
- The performance of RT-PCR assays strongly depends on the amount and quality of sample template RNA. It is strongly recommended to qualify and validate RNA extraction procedures for recovery and purity before testing specimens.
- Suitable nucleic acid extraction systems successfully used in combination with DIAGNOVITAL DETECTION KITS include: Viral NA Isolation Kit from Swabs, bioMérieux NucliSens® systems, QIAamp® Viral RNA Mini Kit, QIAamp® MinElute Virus Spin Kit or RNeasy® Mini Kit (QIAGEN), EZ1 DSP Virus Kit (QIAGEN), Roche MagNA Pure Compact RNA Isolation Kit, Roche MagNA Pure Compact Nucleic Acid Isolation Kit, and Roche MagNA Pure 96 DNA and Viral NA Small Volume Kit, and Invitrogen ChargeSwitch® Total RNA Cell Kit.
- Store and keep residual specimens and extracted nucleic acids at -70°C.
- Only thaw the number of specimen extracts that will be tested in a single day.
- Do not freeze/thaw extracts more than once before testing as each freeze/thaw cycle will decrease the RNA quality.
- It may be possible to use patient samples directly, depending on the sample type. However, this may require a prior lysis step and titration of the amount on sample that can be used without inhibiting the reaction. This procedure has not been validated, use of isolated RNA is recommended.
Reaction Setup
- Make sure that all necessary equipment and devices are suitable, calibrated and functional before starting the experiments.
- Decontaminate equipment and workspace and prepare everything needed for the following experiment to keep the workflow short and repeatable.
- Switch on the PCR detection system and program it to avoid delays after setting up the reactions.
- Thaw all components of DIAGNOVITAL® SARS-CoV-2 Multiplex on ice and mix gently but thoroughly to ensure even distribution of components. Collect liquid at the bottom of the tube with a quick spin (via microcentrifuge).
- Set up your Master mix Plate:
- Always prepare control reactions with nuclease-free dH2O instead of sample material (NTC) to detect contamination in your reagents.
- Always include the assay for the negative Human Extraction Control (HEC) to evaluate the quality of your RNA isolate.
- When using the provided target positive control (TPC), use 4 μl / reaction (max volume of the reaction should be 20 μ).
- > 2 replicates / samples are strongly recommended.
- Prepare enough master mix for all planned reactions. It is recommended to prepare a master mix for 2 additional reactions to compensate for pipetting inaccuracies.
Component Volume 50X VitaScriptTM Reverse Transcriptase 1 µl DIAGNOVITAL® SARS-CoV-2 Multiplex Mix 15 µl isolated sample RNA / TPC /NTC 4 µ l/ 4µ / 4µ dH20 - Distribute the master mix to your strips/plate. An example setup is given in Fig 1.
1 2 3 4 5 6 7 8 9 10 11 12 A Master Mix Master Mix Master Mix Master Mix Master Mix Master Mix Master Mix Master Mix Master Mix Master Mix Master Mix Master Mix B C D E F G H Figure 1: Example pipetting scheme for the distribution of master mixes with the individual assay mixes
- Transfer the Master mix Plate to a separate workspace to add the sample material. Preparing reagents and final reaction setup in separate workspaces helps to avoid contamination of equipment and reagents with sample material.
- Prepare negative reactions first and seal them before handling positive samples. It is recommended to only bring potentially positive sample material and the included target positive control to the workspace once the NTC is prepared and sealed.
- Add your samples to the Master mix Plate. An example setup is given in Fig 2:
1 2 3 4 5 6 7 8 9 10 11 12 A NTC S1 S2 S3 S4 S5 S6 S7 S8 S9 S10 TPC B C D E F G H Figure 2: Example pipetting scheme for the addition of samples. The bottom half of the plate could be used for replicates with an identical setup
- Keep reactions on ice until transferring them to the PCR device.
- Transfer the reactions to the PCR device, then proceed according to these guidelines:
Step Cycles Temperature Duration Reverse Transcription 1 50°C 5 minutes Initial Duration 1 95°C 5 minutes Amplification 40 95°C 5 seconds 60°C* 30 seconds *Enable Data Collection for FAMTM (for virus detection) and HEX/VIC (for human extraction control). If required, set Passive Reference to ROX.
- Once the run is finished, do not open the reaction tubes to avoid contamination and discard according to local guidelines and regulations. Do not autoclave as this may contaminate laboratory equipment with amplicons.
Analysis & Troubleshooting
Exemplary Result
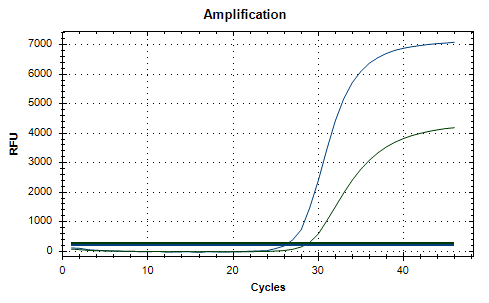
- dH2O controls (NTC) must not give a positive Ct for any assay. If they do, the reaction was contaminated with sample RNA / DNA. Decontaminate equipment and workspace and repeat the reactions.
- For a sample to be considered positive for SARS-CoV-2, the FAM™ channel must give a positive Ct value. Amplification of the HEC in the HEX/VIC channel is expected around Ct < 35 cycles. If the HEC fails to amplify, the sample must still be considered positive.
- For a sample to be considered negative for SARS-CoV-2 , the SARS-CoV-2 assays in the FAM™ channel must not give a positive Ct value. The HEC must give a positive Ct value (< 35 cycles) in the HEX/VIC channel for these samples to ensure that sample material of suitable quality was present.
- All reactions containing RNA isolate must give positive Ct values for the HEC assay. The Ct values should be < 35 cycles. Failure to amplify the negative human extraction control indicates a flawed RNA extraction or loss of RNA isolate due to RNase contamination. The sample is not sufficient, results cannot be interpreted.
- When using the TPC, a positive Ct in the FAM™ channel must be observed. The Ct value for the TPC should be < 35 cycles. If the Ct value does not correspond to the expected value or not all assays are tested positive, PCR was compromised. Check the reaction setup and PCR device settings and repeat the reactions. Repeated freeze and thaw cycles of the TPC can compromise its quality resulting in late Ct values.
- If no amplification signal is observed for any assay, PCR was inhibited. Check reaction setup and device settings and repeat the RNA extraction if necessary. Results are invalid and cannot be interpreted.
| SARS-CoV-2 | HEC | Interpretation |
| + | + | SARS-CoV-2 target sequences & HEC were amplified. The sample is considered positive for SARS-CoV-2. |
| - | + | Only the target sequence for the HEC was amplified. The sample is considered negative for SARS-CoV-2. |
| + | - | SARS-CoV-2 target sequences were amplified but not the HEC. The sample must still be considered positive for SARS-CoV-2. This outcome is possible when having an unusually high virus titer, or the sample was not of human origin, but cell culture derived or analysis of surface contamination. |
| - | - | PCR was inhibited, results are invalid. |
| + | - | Expected result for TPC. |
Limitations
- For reliable results, it is essential to adhere to the guidelines given in this manual. Changes in reaction setup or cycling protocol may lead to failed experiments.
- Depending on the sample matrix, inhibitors may be present in the isolated RNA and disable reverse transcription and/or PCR amplification. If this is the case, another sample type or isolation method may be beneficial.
- Spontaneous mutations within the target sequence may result in failure to detect the target sequence.
- Results must always be interpreted in consideration of all other data gathered from a sample. Interpretation must be performed by personnel trained and experienced with this kind of experiment.
Trademarks
DIAGNOVITAL®, NucliSens® (bioMérieux), QIAamp®, RNeasy® (QIAGEN), ChargeSwitch® (Invitrogen), ROXTM, FAMTM (Life Technologies), DNAZapTM, DNA AwayTM, RNAse AwayTM
Registered names, trademarks, etc. used in this document, even if not specifically marked as such, are not to be considered unprotected by law.
ZQA.IVDM.CV.QOS.00 Revision Date/Revision No: 17.07.2020/00
